- Bergey's Manual Chart
- Bergey's Manual Identification Chart
- Bergey's Manual Flow Chart
- Bergey's Manual Chart Gram Positive Rod
Date
Size
Views
Categories
Bergey's manual of determinative bacteriology by American Society for Microbiology; Bergey, D. (David Hendricks), 1860-1937; Breed, Robert S. (Robert Stanley), 1877-1956. The first edition of Bergey's Manual of Determinative Bacteriology was initiated by action of the Society of American Bacteriologists (now called the American Society for Microbiology) by appointment of an Editorial Board consisting of David H. Bergey (Chairman), Francis C. Harrison, Robert S. Breed, Bernard W. Hammer and Frank M. Bergeys Manual Flow Chart Bergey Manual Of Determinative #4 – Flow Chart to Determine Staphylococcus Aureus, with 42 Similar files. The manual was published subsequent to the Bergey's Manual of Determinative Bacteriology, though the latter is still published as a guide for identifying unknown bacteria. Identification flow charts 1 Bergey’s Manual of Determinative Bacteriology All of the unknowns will fall into the following groups in Bergey's Manual of Determinative Bacteriology (The pink book on the shelf in the laboratory). GROUP 4 Description: Gram Negative, Aerobic/Microaerophilic rods and cocci.
Transcript
Bergey’s Manual of Determinative Bacteriology
All of the unknowns will fall into the following groups in Bergey's Manual of Determinative
Bacteriology (The pink book on the shelf in the laboratory).
GROUP 4
Description: Gram Negative, Aerobic/Microaerophilic rods and cocci
Key differences are: pigments/fluorescent, motility, growth requirements, denitrification,
morphology, and oxidase, read Genera descriptions
Examples: Acinetobacter, Pseudomonas, Beijerinckia, Acetobacter
GROUP 5
Description: Facultatively Anaerobic Gram negative rods
Key differences are: growth factors, morph., gram rxn., oxidase rxn., read Genera descriptions
Examples: Family Enterobacteriaceae and Vibrionaceae
GROUP 17
Description: Gram-Positive Cocci
Key differences are: oxygen requirements, morph., growth requirements (45°C and supplements),
read Genera descriptions
Examples: Micrococcus, Staphylococcus, Streptococcus, Enterococcus, Lactococcus, Aerococcus
GROUP 18
Description: Endospore-Forming Gram positive rods and cocci
Key differences are: oxygen requirements, motility, morph, catalase
Examples: Bacillus, Clostridium
GROUP 19
Description: Regular, Nonsporlating Gram positive rods
Key differences are: morph., oxygen require, catalase
Examples: Lactobacillus, Listeria
GROUP 20
Description: Irregular, Nonsporlating Gram-positive rods
Key differences are: catalase, motility, morph., read Genera descriptions
Examples: Actinomyces, Corynebacterium, Arthrobacter, Propionibacterium
GROUP 21
Description: Weakly Gram-Positive Nonsporlating Acid Fast Slender Rods
Key differences are: acid fast, growth
Examples: Mycobacterium
1
Identification flow charts
Differentiation via Gram stains and cell morphology.
Gram Stain & Morphological
Flowchart
Some Examples
Gram Positive
Cocci Bacilli
Filamentous
Round in clusters Oval shape in Club-shaped and/or in Spore-bearing,
& tetrads: Chains: palisades: large, uniform:
Staphylococcus Streptococcus Corynebacterium Bacillus
Micrococcus Peptostreptococcus Listeria Clostridium Extensive Branching rudi-
Peptococcus Enterococcus Erysipelothrix branching: mentry or absent:
Mycobacterium (Acid Fast) Actinomyces Erysipelothrix
Propionobacterium Arachnia Lactobacillus
Nocardia (partially Eubacterium
acid-fast)
Streptomyces
Gram Negative
Cocci Bacilli
Neisseria Medium-sized Tiny-cocco- Pleomorphic Exaggerated Curved, Uniformly
Veillonella cocco-bacillary: bacillary: coccobacillary pointed ends: comma-shaped: Bacillary:
Acinetobacter Brucella & filamentous: Fusobacterium Vibrio Enterobacteriaceae
Moraxella Bordetella Haemophilus nucleatum Campylobacter Pseudomonas
Bacteroides Bacteroides Spirillium Aeromonas
Pasteurella Alcaligenes
Francisella Chromobacterium
Actinobacillus Coiled &
Eikenella sphero-plastic:
Cardiobacterium Streptobacillus
Flavobacterium Fusobacterium
2
Identification flow charts
Gram Positive Rods ID Flowchart
Gram Positive Rods
Bacillus spp.
Clostrididium spp.
Corynebacterium spp.
Lactobacillus spp.
Mycobacterium spp.
Mycobacterium smegmatis
Spore Forming
+ - +
Bacillus spp. Corynebacterium spp.
Clostridium spp Lactobacillus spp. Acid Fast
Mycobacterium spp.
-
Strict Anaerobes Corynebacterium spp
Lactobacillus spp.
+ -
Clostridium spp. Bacillus spp.
You sure you have this? See separate flowchart.
Go to Bergey's. Catalase
+ -
Corynebacterium spp. Lactobacillus spp.
Starch Hydrolysis
Glucose Ferm. Activity
+ -
Acid & Gas Acid
Corynebacterium kutsceri Corynebacterium xerosis
Lactobacillus fermenti Lactobacillus casei
Lactobacillus delbrueckii
Mannitol
+ -
Lactobacillus casei Lactobacillus delbrueckii
3
Identification flow charts
Bacillus spp. ID Flowchart
Bacillus spp.
Starch Hydrolysis B. pumilus
(amylase) (VP is pos., rest are neg.)
B. subtilis
B. marinus
B. cereus - B. sphaericus
B. azotoformans
B. megaterium B. larvae
B. stearothermophilus B. schlegelii B. pasteurii
B. polymyxa
+ (grows at 55°C, rest no) (round spore, rest
B. popilliae are oval)
B. mycoides VP B. pasteurii B. popilliae
B. macerans (add creatine as catalyst B. lentimorbus
B. thuringiensis w/ reagents) -
B. azotoformans
B. macquariensis
B. badius
B. licheniformis
B. insolitus
B. lentus + - B. larvae
B. alvei
B. subtilis B. megaterium B. laterosporus Catalase Citrate
B. anthracis
B. alcalophilus B. cereus B. stearothermophilus
B. coagulans B. polymyxa B. macerans
B. mycoides B. pantothenticus + +
B. brevis
B. macquariensis B. badius -
B. circulans B. thuringiensis
B. licheniformis B. lentus (spore is not
B. firmus round, rest are) B. azotoformans B. larvae
B. pantothenticus B. alvei B. alcalophilus (gelatinase pos.)
B. badius B. insolitus
B. anthracis B. popilliae
B. brevis B. laterosporus
B. coagulans (gelatinase neg.)
B. circulans B. pasteurii
B. marinus
B. sphaericus
Cell Diamenter ! 1'm Swollen Cell Swollen Cell
(the width) (containing spore) (containing spore)
+ - + -
B. anthracis B. subtilis B. stearothermophilus B. megaterium + -
(non-motile) B. polymyxa (growth at 55°C-rest neg.) (citrate pos.)
B. thuringiensis B. lichenifomis B. macerans B. badius B. sphaericus B. insolitus
(makes insecticidal B. alvei B. pantothenticus (citrate neg.) B. pasteurii (acid from glucose)
protein)
B. coagulans B. macquariensis B. laterosporus B. marinus
B. mycoides B. lentus (oval spore, (acid from glucose)
(colony has a (catalase neg.-rest pos.) rest are round)
rhizoidal appearance) B. alcalophilus
B. cereus B. brevis
(Motile - It is this B. circulans
one ) Nitrate Reduction
Citrate + -
6.5% NaCl Growth B. pasteurii B. sphaericus
+ -
B. coagulans B. polymyxa + -
B. licheniformis (manitol pos.)
B. subtilis B. alvei B. macerans B. pantothenticus
(manitol neg.) B. macquariensis (acid via arabinose neg.)
B. alcalophilus B. circulans
B. brevis (acid via arabinose pos.)
B. circulans
6.5% NaCl Growth
B. macerans
+ - Acid from Arabinose
+ (gas from glucose pos.)
B. alcalophilus
B. subtilis B. coagulans - (gas from glucose neg.,
(no growth at 55°C) no growth < pH 7.0)
B. licheniformis B. macquariensis B. circulans
(growth at 55°C) (methyl red pos.) (gas from glucose neg.,
B. brevis growth < pH 7.0)
(methyl red neg.)
4
Identification flow charts
Gram Positive Cocci ID Flowchart
Gram Positive Cocci
Streptococcus spp.
Micrococcus spp. - (see Bergey’s)
Staphylococcus spp.
Streptococcus spp. Bile Esculin
Enterococcus spp. (growth with esculin hydrolysis) Enterococcus faecalis
Enterococcus faecium
Enterococcus hirae
+ Enterococcus mundtii
Catalase -
Streptococcus pneumoniae
Streptococcus mitis
Streptococcus pyogenes Group A
+ Streptococcus spp. Group B
Enterococcus spp. Enterococcus spp.
Micrococcus spp. Streptococcus spp. Streptococcus spp.
Staphylococcus spp.
Growth w/ Tellurite
Gamma (!)
Mannitol Fermentation
+ -
- Hemolysis
+ Enterococcus faecalis Enterococcus faecium
Staphylococcus aureus Staphylococcus spp. (mannitol pos., white colony)
Micrococcus spp. Enterococcus hirae
(mannitol neg.)
Enterococcus mundtii
(mannitol pos., yellow colony)
Beta (')
Yellow Pigment (Colony)
Alpha (#)
Streptococcus pyogenes Group A
+ - Streptococcus spp. Group B
Micrococcus spp. Staphylococcus spp.
Pyrrolidonearylamidase
(PYR Test)
Glucose Fermentation Novobiocin Sensitivity Streptococcus pneumoniae
Streptococcus mitis
+
Micrococcus varians Yes +
Streptococcus pyogenes Group A
Staphylococcus epidermidis -
- - No Streptococcus spp. Group B
(Streptococcus agalactiae)
Micrococcus luteus Staphylococcus saprophyticus
Optochin Sensitivity
Yes No
Streptococcus pneumoniae Streptococcus mitis
5
Identification flow charts
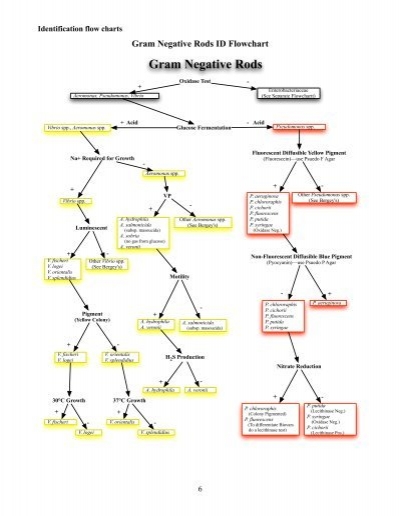
Gram Negative Rods ID Flowchart
Gram Negative Rods
Oxidase Test -
+
Enterobacteriaceae
Aeromonas, Pseudomonas, Vibrio (See Separate Flowchartt)
+ Acid - Acid
Vibrio spp., Aeromonas spp. Glucose Fermentation Pseudomonas spp.
Fluorescent Diffusible Yellow Pigment
Na+ Required for Growth (Fluorescein)—use Psuedo F Agar
-
Aeromonas spp.
+ -
+
VP P. aeruginosa Other Pseudomonas spp.
Vibrio spp. P. chlororaphis (See Bergey's)
- P. cichorii
+
P. fluorescens
A. hydrophila Other Aeromonas spp. P. putida
A. salmonicida (See Bergey's) P. syringae
Luminescent (subsp. masoucida) (Oxidase Neg.)
A. sobria
(no gas from glucose)
A. veronii
+ -
Non-Fluorescent Diffusible Blue Pigment
V. fischeri Other Vibrio spp. (Pyocyanin)—use Psuedo P Agar
V. logei (See Bergey's)
V. orientalis
V. splendidius Motility
- +
P. chlororaphis P. aeruginosa
- P. cichorii
Pigment + P. fluorescens
(Yellow Colony) A. hydrophila P. putida
A. salmonicida
A. veronii (subsp. masoucida) P. syringae
+ -
V. fischeri V. orientalis
V. logei V. splendidius
H2S Production
Nitrate Reduction
+ -
A. hydrophila A. veronii
+ -
30°C Growth 37°C Growth
P. putida
P. chlororaphis
+ + (Colony Pigmented)
(Lecithinase Neg.)
P. syringae
V. fischeri V. orientalis P. fluorescens (Oxidase Neg.)
- - (To differentiate Biovars
do a lecithinase test)
P. cichorii
V. logei V. splendidius (Lecithinase Pos.)
6
Identification flow charts
Family Enterobacteriaceae Lactose Positive ID Flowchart
Family Enterobacteriaceae
Lactose Fermentation
+ -
Citrobacter diversus Erwinia chrysanthemi See Lactose Negative Flowchart
Citrobacter freundii Escherichia coli for Enterobacteriaceae
Enterobacter aerogenes Klebsiella oxytoca
Enterobacter cloacae Klebsiella pneumoniae
Enterobacter amnigenus (subsp. ozaenae and pneumoniae)
Enterobacter intermedius Serratia fonticola
Erwinia carotovora Serratia rubidaea
Indole
-
The Rest
+
Citrobacter diversus
Escherichia coli
Erwinia chrysanthemi
MR-VP
Klebsiella oxytoca +/+ -/+ Klebsiella pneumoniae
(subsp. pneumoniae)
Enterobacter intermedius Enterobacter aerogenes
Enterobacter cloacae
Enterobacter amnigenus
+/- Erwinia carotovora
Citrobacter freundii Serratia rubidaea
Citrate Serratia fonticola
Klebsiella pneumoniae
(subsp. ozaenae)
Lysine Decarboxylase
+ -
+ -
Klebsiella pneumoniae subsp. pneumoniae Enterobacter cloacae
Citrobacter diversus Escherichia coli Non-motile, No Pigment Acid from sorbitol
(VP neg.) Serratia rubidaea Enterobacter amnigenus
Erwinia chrysanthemi Motile, Red pigment No acid from sorbitol
(VP and H2S pos.) Enterobacter aerogenes
Klebsiella oxytoca H2S Production Motile, No pigment
(VP pos and H2S neg.)
??
Erwinia carotovora
Lysine unknown but Gelatinase
positive; none above are positive.
+ -
Citrobacter freundii Serratia fonticola
Motility pos.
Klebsiella pneumoniae subsp. ozaenae
Motility neg.
7
Identification flow charts
Family Enterobacteriaceae Lactose Negative ID Flowchart
Family Enterobacteriaceae Continued
Lactose Negative Flow Chart
Erwinia cacticida
Proteus mirabilis
Edwardsiella tarda Proteus penneri
Erwinia cacticida Serratia marcescens Salmonella bongori
Morganella morganii Serratia liquefaciens Salmonella enterica
Proteus mirabilis Shigella spp. (boydii, dysenteriae, flexneri) Serratia marcescens
Proteus penneri Shigella sonnei Serratia liquefaciens
Proteus vulgaris Yersinia enterocolitica Shigella sonnei
Providencia stuartii Yersinia pestis - Yersinia pestis
Salmonella bongori Yersinia pseudotuberculosis Yersinia pseudotuberculosis
Salmonella enterica
+ Indole Test
Edwardsiella tarda
Morganella morganii
Urease
Proteus vulgaris
Providencia stuartii ?? + -
Please See Bergey’s to ID: Erwinia cacticida
Shigella spp. Proteus mirabilis
(boydii, dysenteriae, flexneri) Ornithine decarb. pos. Salmonella bongori
Yersinia enterocolitica Proteus penneri Salmonella enterica
Ornithine decarb. neg., motile Serratia marcescens
H2S Yersinia pseudotuberculosis Serratia liquefaciens
Ornithine decarb. neg., non-motile Shigella sonnei
Yersinia pestis
+ -
Edwardsiella tarda Morganella morganii
Motility
Proteus vulgaris Providencia stuartii
+ -
Erwinia cacticida Shigella sonnei
(lysine decarb. neg., the rest pos.) Yersinia pestis
Urease Ornithine Decarboxylase Salmonella bongori
Salmonella choleraesuis
Serratia marcescens
Serratia liquefaciens
+ +
Proteus vulgaris Providencia stuartii
Ornithine Decarboxylase
- -
Edwardsiella tarda Morganella morganii
H2S
+
Shigella sonnei -
+ -
Yersinia pestis
Salmonella bongori Serratia marcescens
KCN growth pos. Red pigment
Salmonella enterica Serratia liquefaciens
KCN growth neg. No Red pigment
(It is this one)
8
The following points highlight the top four Volumes of Bergey’s Manual of Systematic Bacteriology.
Bergey’s Manual of Systematic Bacteriology # Volume I:
Section 1: The spirochetes
Bergey's Manual Chart
Order I. Spirochaetals.
ADVERTISEMENTS:
Family I. Spirochaetaceae e.g., Spirochaeta.
Family II. Leptospiraceae e.g., Leptospira.
Section 2: Aerobic, microaerophilic, motile, helical, vibroid, Gram negative curved bacteria, e.g., Aquaspirlllum, Azospirillum, spirillum .
Section 3: Normotile (or rarely motile), Gram negative curved bacteria.
Family I: Spirosomaceae, e.g., spirosoma.
Section 4: Gram negative aerobic rods and cocci.
Family I: Pseudomonadaceae e.g., Pseudomonas.
Family II: Azotobacteraceae e.g., Azotobacter.
Family III: Rhizobiaceae e.g., Rhizobium.
Family IV : Methylococcaceae e.g., Methylococcus.
Family V : Halobacteriaceae e.g., Halobacterium.
Family VI: Acetobacteriaceae e.g., Acetobacter.
Family VII: Legionellaceae e.g., Legionella.
ADVERTISEMENTS:
Family VIII: Neisseriaceae e.g., Neisseria, Beizeriuckia.
Section 5: Facultatively anaerobic Gram-negative rods e.g.,
Family I: Enterobacteriaceae e.g., Escherichia, shigella, salmonella, klbsiella, Yersia.
Family II: Vibrionaceae, e.g., Vibrio.
Family III: Pasteuellaceae e.g., Actinobacillus Haemophilus.
Section 6: Anaerobic Gram negative straight, curved and helical rods.
Family I: Bacteroidaceae e.g., Bacteroides.
Section 7: dissimilatory sulphate or sulphur reducing bacteria e.g., Desulfuromonas, Desulfobacter.
ADVERTISEMENTS:
Section 8 : Anaerobic Gram-negative cocci.
Family I: Veillonellacae e.g.,veillonella.
Section 9: Rickettsias and chlamydia orders
Order I: Rickettsiales.
Family I: Rickettsiaceae e.g., Rickettsia.
Family II: Bartonellaceae e.g., Bartonella.
Family III: Anaplasmtaceae e.g., Anaplasma
Section 10: Order I: Mycoplasmatales
Family I: Mycoplasmataceae e.g., Mycoplasma, ureoplasma.
Family II: Acholeplasmataceae e.g., Acholeplasma.
ADVERTISEMENTS:
Family III: Spiroplasmataceae e.g., spiroplasma.
Section 11: Endosymbiont.
A . Endosymbiont of Protozoa, Ciliates, Flagellates, amoebae.
B . Endosymbiont of insects .
C . Endosymbiont of fungi and invertebrates other than arthropods.
Bergey’s Manual of Systematic Bacteriology # Volume II:
Section 12: Gram positive cocci.
Family I: Micrococcaceae e.g., Micrococcus
Family II: Deinococcaceae e.g., Deinococcus.
Section 13: Endospore forming Gram-positive rods and cocci e.g., Bacillus, Clostridium.
Section 14: Regular, non-sporing, Gram positive rods e.g., Lactobacillus, Renibacterium.
Section 15: Irregular, non-sporing, Gram-positive rod, e.g., corynebacterium, Microbacterium.
Section 16: The Mycobacteria.
Family: Mycobacteriaceae e.g., Mycobacterium
Section 17: Nocardioforms e.g., Nocardia, rhodococcus.
Bergey’s Manual of Systematic Bacteriology # Volume III:
Section 18: Anoxygenic phototrophic bacteria.
(I) Purple bacteria
Family I: Chromatiaceae e.g., chromatium .
Family II: Ectothiorhodospiraceae, Ectothiorhodospira.
(II) Purple Non sulphur bacteria Rhodospirillum, Rodobacter.
(III) Green Bacteria:
Green sulphur bacteria e.g., chlorobium, chloroberpeptone.
(IV) Multicellular, Filamentous, green bacteria e.g., Chloroflexus, heliothrix.
(V) Genera Incertae sedis Heliobacterium, Erytherobacter.
Section 19: Oxygenic photosynthetic bacteria Cyanobacteria.
Family I: Chrococcaceae.
Family II: Pleurocapraceae.
Family III: Oscillatoriaceae e.g., spirulina, oscillatoria
Family IV : Nostocaceae e.g., Anabena, Nostoc.
Family V: Stigonemataceae e.g., Chlorogloeopsis.
B: Prochloraceae e.g., Prochloron.
Section 20: Aerobic Chemolitholrophic bacteria and associated organisms.
A. Nitriflying bacteria.
Family: Nitrobaacteriaceae e.g., Nitrobacter, Nitrococcus, Nitrosomonas.
B. Colourless Sulphur bacteria, e.g., Thiobacterium, Macromonas, Thiospira.
C. Obligately chemolithtrophic, Hydrogen bacteria e.g., Hydrogenobacter.
D. Iron and manganese oxidizing and/or depositing bacteria e.g.,
Family: Siderocapsaceae e.g., Siderocapsa.
E. Magnotactic bacteria, e.g., Aquaspirillum, Maganotacticum.
Section 21: Budding and/or Appendaged bacteria
1. Prosthecate Bacteria.
A. Budding Bacteria, Genus, Hyphomonas, Prosthecomicrobium.
B. Non budding bacteria, Caulobacter, Prosthecobacter.
2. Nonprosthecate bacteria.
A. Budding bacteria lack peptidoglycan, planctomyces, contain peptidoglycan e.g., Blastobacter.
B. Non-budding bacteria e.g., Galionella, Nevskia.
Section 22: Sheathed Bacteria, e.g., Sphaerotilus, Leptothrix, clonothrix.
Section 23: Nnphotsynthetic, non-fruiting Gliding bacteiua.
Family I: Cytophagaceae, e.g., cytophaga.
Family II: Lysobacteriaceae e.g., Lysobacter.
Family III: Beggiatoaceae e.g., Beggiatoa, Thiothrix, Thioploca.
Other families:
Family: Simonriellaceae e.g., Simonsiella.
Family: Pelonemataceae, e.g., pelonema.
Section 24: Fruiting Gliding bacteria (Myxobacteria).
Family I: Myxococcaceae e.g., Myxococcus.
Family II: Archangiaceae e.g., Archangium.
Family III: Cystobacteriaceae e.g., Cystobacter.
Bergey's Manual Identification Chart
Family IV : Polyangiaceae e.g., Polyangium.
Section 25 : Archaebacteria.
Group I : Methanogenic archae bacteria.
Family I: Methanobacteraceae e.g., Methanobacterium.
Family II: Methanothermaceae e.g., Methanthermus.
Family III: Methanomicrobiaceae e.g., Methanomicrobium.
Family IV : Methanosarcinaceae e.g., Methanosarcina.
Family V : Methanplanaceae e.g., methanoplanus.
Group II : sulphate reducer Archace bacteria.
Family : Archaeoglobaceae e.g., Archaeoglobus.
Group III: Extrenely halophilic Archaebacteria.
Bergey's Manual Flow Chart
Family : Halobacteriaceae e.g., Halobacterium.
Group IV : Cell wall less archaebacteria.
Family: Thermoplasmaceae.
Group V: Extremely thermophilic sulphate metabolizers. Family: Thermococcaceae e.g.,Thermocccus.
Bergey’s Manual of Systematic Bacteriology # Volume IV:
Section 26: Nocardioform actinomeycetes e.g., Nocardia, Rhodococcus.
Section 27: Actinomycetes with multi-locualar sporangia e.g.,. Frankia, Dermatophilus.
Section 28: Actinoplanetes e.g., Actonoplanes, Micromonospora.
Section 29: Streptomycetes and related genera a e.g., streptomyces, Kineosporia.
Section 30: Maduromycetes, e.g., actinomadura, Streptosporangium.
Section 31: Thermomonospora and related genera e.g., Nocardiopsis.
Section 32: Thermactinomycetes e.g., Thermoactimoneyces.
Section 33: Other genera e.g., Glycmyces, Pasteuria, Saccharothrin.